How to input knowledge to the Machines (aka ) Machine learning ?!¶
Learning Algorithms :¶
Task T :¶
* Classification
* Regression
* Transcription
* Machine translation
* Anomaly detection
* Synthesis and sampling
* Imputation of missing values
* DenoisingThe Performance Measure P :¶
* Accuracy
* Precision, Recall
* Reconstruction Error
* RMSE ,MSE ,MAEThe Experience E :¶
* Supervised Learning p(y/x)
* Unsupervised Learning p(x)
* Reinforcement learning (continous learning)
* Small line which separates Supervised from unsupervised learning (Arinthaal ... arinthaal....)
Hint¶
Generally ,The Best practice is to choose the Dataset/design matrix for your algorithm in suchway that your model able to see all possible combinations of examples (data points)¶
What you want from Learning Algorithms !?¶
* Generlization What are all the learning Algorithms you know ?¶
we will broadly classify the Algorithms into three parts : (for simplicity)¶
* Statistical models
* Linear Models
* Tree based Models
* Neural NetworksDrawbacks of Linear Models¶
* Scaling is required
* Normalization is required
* Encoding categorical varaibles is challenging
* Interpreting the results needs some addtional work
* Implementing Multi class classification needs some extra thinking
* Rest are Assignment !!!!DECISION TREE¶
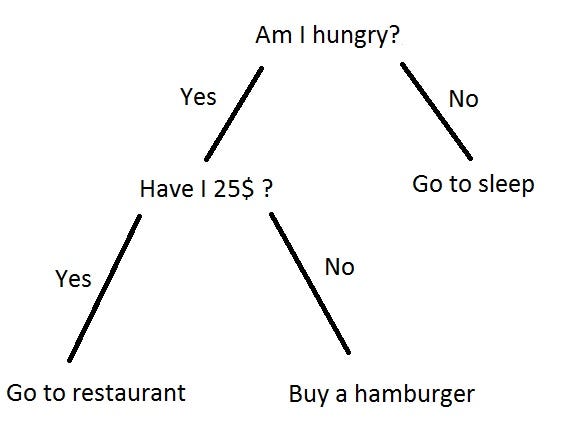
Example of Decsion tree¶
think about how to get predictions ?¶
So Which feature to Query and which threshold to choose !? how to write a generlized code for best split !???¶
Consider the following example to split red from green
which one is good split !?¶
The Basic Algorithm¶
1 Start at the root node as parent node
2 Split the parent node at the feature xi to minimize the sum of the child node impurities (maximize information gain)
3 Assign training samples to new child nodes
4 Stop if leave nodes are pure or early stopping criteria is satisfied, else repeat steps 1 and 2 for each new child node
Stopping Rules¶
1 The leaf nodes are pure
2 A maximal node depth is reached
3 Splitting a note does not lead to an information gainTalking about what to reduce (i.e how impurity has to be reduced) we have following measurements¶
* Gini index
* Entropy
* Misclassification Error
* ID3
* Chi-Square
* Reduction in Variance
Reference :¶
https://medium.com/@rishabhjain_22692/decision-trees-it-begins-here-93ff54ef134
https://www.bogotobogo.com/python/scikit-learn/scikt_machine_learning_Decision_Tree_Learning_Informatioin_Gain_IG_Impurity_Entropy_Gini_Classification_Error.php
https://sebastianraschka.com/faq/docs/decisiontree-error-vs-entropy.htmlInformation gain¶
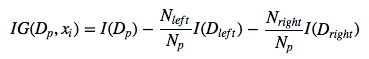
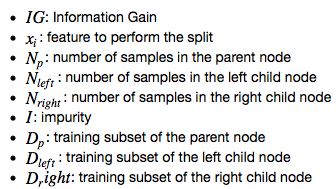
Gini index :¶
Gini index (a criterion to minimize the probability of misclassification)
Gini =$1-\sum_jp_j^2$
where $p_j$ is the probability of class j.
More Generalized Form¶
Entropy :¶
* Way to measure impurity
* calculates the homogeneity of a sample. If the sample is completely homogeneous the entropy is zero and if the sample is equally divided then it has entropy of one
Entropy = $-\sum_jp_j\log_2p_j$
Example :¶
entropy = $-1 \log_2 1 = 0$
entropy = $-0.5 \log_2 0.5 - 0.5 \log_2 0.5 = 1$
In [ ]:
In [1]:
from sklearn.datasets import load_diabetes
from sklearn.tree import DecisionTreeRegressor,DecisionTreeClassifier
from sklearn.model_selection import train_test_split
X,y=load_diabetes(return_X_y=True)
In [2]:
X_train,X_test,y_train,y_test=train_test_split(X,y)
X_train.shape,y_train.shape
Out[2]:
((331, 10), (331,))
In [3]:
import matplotlib.pyplot as plt
import numpy as np
plt.plot(y)
plt.show()
In [4]:
dtr=DecisionTreeRegressor()
dtr.fit(X_train,y_train)
Out[4]:
DecisionTreeRegressor(criterion='mse', max_depth=None, max_features=None,
max_leaf_nodes=None, min_impurity_decrease=0.0,
min_impurity_split=None, min_samples_leaf=1,
min_samples_split=2, min_weight_fraction_leaf=0.0,
presort=False, random_state=None, splitter='best')
In [5]:
pred=dtr.predict(X_test)
plt.plot(pred,label='prediction')
plt.plot(y_test,label='Actual')
plt.legend()
plt.show()
In [6]:
from sklearn.metrics import mean_squared_error,mean_absolute_error
mae=mean_absolute_error(y_test,pred)
mse=mean_squared_error(y_test,pred)
rmse=mse**0.5
plt.bar(['mae','mse','rmse'],[mae,mse,rmse])
plt.title('mae={},mse={},rmse={}'.format(mae,mse,rmse))
plt.show()
In [1]:
!pip install pydotplus
Requirement already satisfied: pydotplus in c:\users\gurunath.lv\appdata\local\continuum\anaconda3\lib\site-packages (2.0.2) Requirement already satisfied: pyparsing>=2.0.1 in c:\users\gurunath.lv\appdata\local\continuum\anaconda3\lib\site-packages (from pydotplus) (2.2.0)
In [11]:
from treeinterpreter import treeinterpreter as ti
prediction, bias, contributions=ti.predict(dtr,X_test)
In [14]:
import pydotplus
from sklearn.tree import export_graphviz
from IPython.display import Image,HTML,SVG
from io import StringIO
dot_data = StringIO()
export_graphviz(dtr, out_file=dot_data,
filled=True, rounded=True,
special_characters=True)
graph = pydotplus.graph_from_dot_data(dot_data.getvalue())
#Image(graph.create_png())
In [20]:
conda_fix(graph)
Image(graph.create_jpg())
Out[20]:
Advantages of Tree based Models :¶
* Easy categorical Handling
* interpreting the results are easy
* No scaling /Normalization are required
* Rest are Assignment !!!